Ghidra Scripting for Analysis and Machine Learning Applications
by Coleman Kane
Table of Contents
- Enumerating the Instruction at an Address
- Statistical Instruction Analysis
- Using Ghidra CLI
- Accessing Ghidra P-Code
The prior exercise had you use Ghidra to perform some post-processing work on a program being analyzed. In this section, we will explore some uses of Ghidra for the purpose of pulling insights out of functions or whole programs. This approaches frequently have applications across n a range of statistical analyses, including machine learning, as well as other places.
Enumerating the Instruction at an Address
From the prior examples, we learned that there’s a variable accessible to all of your Ghidra scripts
named currentAddress which stores the address of the current cursor location within the Listing
View. Furthermore, there’s a member function to your script named getFunctionContaining() that
accepts an address, and provides the data structure representing a single function in Ghidra, that
contains the provided address.
fn = getFunctionContaining(currentAddress)
In the above, the following data types are being employed:
currentAddress: A ghidra.program.model.address.Addressfn: A ghidra.program.model.listing.Function
Ghidra offers a convenient function named getFirstInstruction() that can optionally take a Function argument, and return an
Instruction class that represents the first executable instruction in a function. To get the first instruction of the function
into a variable where you can use it:
first_instr = getFirstInstruction(fn)
In the above, first_instr is an instance of type:
There are a number of really interesting functions in both of these that provide some insights:
getMnemonicString(): Gets a string containing the instruction operand name that Ghidra displaysgetBytes(): Get an array of the bytes comprising this instructiongetComment(): Get the text of one the comments that may have been addedgetReferencesFrom(): Get information about what code/data this instruction is referencinggetReferenceIteratorTo(): Get an iterator yielding all of the other references that point to this instructiongetPcode(): Get the Ghidra P-Code representation of the instructiongetPrevious()/getNext(): Return the prior or next instruction, allowing you to move forward or backward
To start, we could write a short script like the following to dump out the first instruction opcode in the selected function, called GetFirstInstructionFromFunction.py
# Retrieves and displays the summary counts for instructions used within
# a function.
#
# @category CS6038.Demo
#
# Import some of the Ghidra classes we will be using
from ghidra.util.task import ConsoleTaskMonitor
# Get the function where the cursor is presently located
fn = getFunctionContaining(currentAddress)
# Identify where the first instruction is within the function
instr = getFirstInstruction(fn)
# Print the mnemonic (the text name) of the opcode to the console
print(instr.getMnemonicString())
And, running it against a randomly chosen function should yield output similar to below. Your mileage will vary depending upon which function you choose.
GetFirstInstructionFromFunction.py> Running...
PUSH
GetFirstInstructionFromFunction.py> Finished!
Statistical Instruction Analysis
This may not be super useful on its own, but let’s try something more complex (and potentially more useful). Say we are interested in knowing some information about the instruction composition of a particular function. It might be useful to count each of the instructions, and then report this summary table of frequency counts, for the function. To consider this problem however requires us to consider some programmatic possibilities.
- The entry-point into a function may not necessarily have to be the instruction with the lowest/earliest address
- While blocks within functions have to be contiguous, any branches to other blocks don’t necessarily need to be “compact”, or adjacent.
- Functions can be declared with other functions embedded partially or wholly within them, as long as the execution path accounts for this
With the above constraints in mind, it is important to inspect the API for the Function type, as well as its parent type,
FunctionDB.
Using the above documentation, a few useful functions stand out for addressing the problem of figuring out which instructions are part of a function, as well as the upper and lower address bounds of that function (our search space).
getBody(): Returns anAddressSetViewthat provides an interface to a set of address ranges for the blocks of the function.getEntryPoint(): Returns the enry point to this function (the address of the first instruction executed when it is called)
Within the ghidra.program.model.address.AddressSetView
type, returned by getBody(), are some additonal helpful functions:
contains(): Returnstrue/falsedepending upon if the encapsulated address ranges contain the provided address(es) or address range(s)getMinAddress(): Provides the lowest numerical address contained within this setgetMaxAddress(): Provides the highest numerical address contained within this set
The AddressSetView represents an immutable collection of address ranges. As was mentioned earlier, each of the contiguous blocks within
a function must be contiguous within the compiled program, but there are minimal requirements for the blocks themselves to also be
adjacent within the program. This is largely done out of convention, as most CPUs can optimize execution better when related code is
adjacent, as well as to aid in debugging and analysis. Though not extremely common, you will likely encounter functions that exhibit
this behavior of having their blocks non-adjacent, so learning about this possibility, and considering it into your designs, will
be useful. There are some tools out there that exist to help malware developers spread around different blocks of functions for the
purpose of making manual analysis more difficult.
Taking this into consideration, to generate a statistical summary of a function’s instructions, I will want to structure the search in the following manner:
- Find the earliest (lowest) memory address to start looking
- Walk through eacho of the Instructions using the
Instruction.getNext()method - Using the result of
Function.getBody(),AddressSetView.contains()can confirm for us that an instruction is within a function - We can use the result of
contains()to determine whether we count an instruction or skip over it
Using the above instructions, we can extend our earlier script to build an instruction-based statistical analyzer, named GetFunctionInstructions.py:
# Retrieves and displays the summary counts for instructions used within
# a function.
#
# @category CS6038.Demo
#
# Use the JSON library for output
import json
# Import some of the Ghidra classes we will be using
from ghidra.util.task import ConsoleTaskMonitor
# Get the function where the cursor is presently located
fn = getFunctionContaining(currentAddress)
# Use AddressSetView.getMinAddress() to get the location of the earliest fragment of the function
instr = getInstructionAt(fn.getBody().getMinAddress())
# We want to store the results in a Python dict data structure. Each key will have an instruction
# mnemonic, while each value will be the counts, with absent instructions representing zero counts
instr_map = {}
# Make sure that the instruction we're analyzing is not outside of the function's max memory range
while instr.getMinAddress() <= fn.getBody().getMaxAddress():
# If the instruction is contained by one of the fragments represented in the AddressSetView, then
# it is part of the code for the function, and we should count it
if fn.getBody().contains(instr.getMinAddress()):
# Get the string mnemonic name from the instruction
opcode = instr.getMnemonicString()
# If an entry exists in the dict, then increment its counter, otherwise, create a new entry
# populated with 1
if opcode in instr_map:
instr_map[opcode] += 1
else:
instr_map[opcode] = 1
# Advance the cursor to the next instruction
instr = instr.getNext()
print(json.dumps(instr_map, sort_keys=True, indent=2))
# The below line will be useful for saving the output to a file handle for machine processing
# print(json.dumps(instr_map))
The above yields the following output in the console:
GetFunctionInstructions.py> Running...
{
"CALL": 6,
"CMP": 2,
"INC": 1,
"JMP": 1,
"JNZ": 1,
"JZ": 1,
"LEA": 2,
"LEAVE": 1,
"MOV": 5,
"POP": 3,
"PUSH": 22,
"RET": 1,
"SUB": 1,
"XOR": 1
}
GetFunctionInstructions.py> Finished!
Using Ghidra CLI
In addition to the GUI Ghidra utility, which is extremely powerful, Ghidra also offers a command-line utility
named analyzeHeadless, that can be used to run scripts as well. This is extremely helpful for automating bulk
analysis across 1,000’s of artifacts, or handling long-running computational analysis on a server where you may
not want to deal with setting up a GUI. In any Ghidra package, it is located in the support/ subdirectory.
To start, we will recall that in the previous module two scripts were written, HelloScript.py and
HelloScript.java. As well, it is important to review a bit about Ghidra’s project & directory structure.
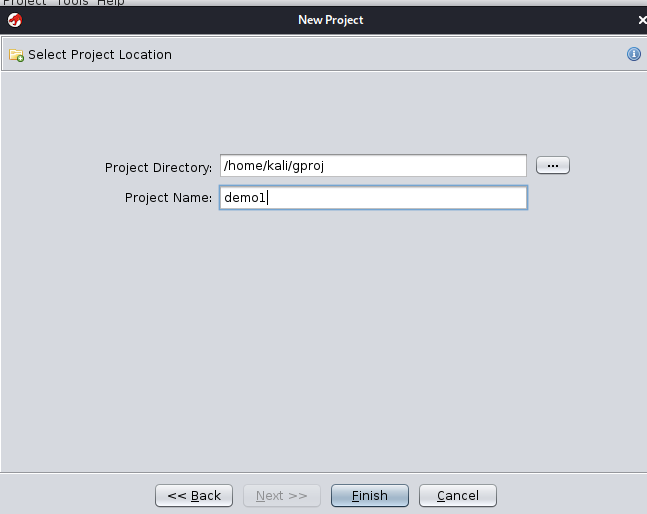
When creating a new project, you tell Ghidra where your project directory is going to be, as well as the name of
the new project to create in that folder. The project will have a file ending in *.gpr and a directory ending
in *.rep created in the folder. This enables the reuse of the same Project Folder to store multiple Ghidra
projects, which can be helpful for organization. Additionally, while Ghidra is running, some .lock and .lock~
files will be created, in order to allow Ghidra to protect the project from being opened more than once from the
same file (which is not supported).
After creating the above project, the gproj/ folder will contain demo1.gpr and demo1.rep/. Upon creating
the new project, use “I” to import the algo1 program from the earlier examples, and then open it so that the
Ghidra analyzer will perform analysis on the program. Save it, and then exit Ghidra.
- It is important to remember that you cannot use
analyzeHeadlesson a project while it is also open in Ghidra
The usage from analyzeHeadless -help:
Headless Analyzer Usage: analyzeHeadless
<project_location> <project_name>[/<folder_path>]
| ghidra://<server>[:<port>]/<repository_name>[/<folder_path>]
[[-import [<directory>|<file>]+] | [-process [<project_file>]]]
[-preScript <ScriptName>]
[-postScript <ScriptName>]
[-scriptPath "<path1>[;<path2>...]"]
[-propertiesPath "<path1>[;<path2>...]"]
[-scriptlog <path to script log file>]
[-log <path to log file>]
[-overwrite]
[-recursive]
[-readOnly]
[-deleteProject]
[-noanalysis]
[-processor <languageID>]
[-cspec <compilerSpecID>]
[-analysisTimeoutPerFile <timeout in seconds>]
[-keystore <KeystorePath>]
[-connect <userID>]
[-p]
[-commit ["<comment>"]]
[-okToDelete]
[-max-cpu <max cpu cores to use>]
[-loader <desired loader name>]
- All uses of $GHIDRA_HOME or $USER_HOME in script path must be preceded by '\'
Here’s an attempt using the demo1 project in the above screenshot, after importing and analyzing algo1. Note
that I add the -noanalysis argument to turn off the auto-analysis code, which already ran when we imported the
file using the GUI, so execution will speed up (somewhat) if we tell it to skip re-doing this step. By default,
analyzeHeadless will also re-run the Ghidra analyzers, to update the analysis database with whatever the latest
Ghidra version opening the file is.
/opt/ghidra_9.1.1_PUBLIC/support/analyzeHeadless ~/gproj/ demo1 \
-process algo1 \
-noanalysis \
-postScript ~/cs6038_ghidra_scripts/HelloScript.py
The above command tells Ghidra to open the demo1 project from inside of the ~/gproj/ project folder, and then
process the imported artifact algo1. The processing will skip the analysis phase because of the -noanalysis
argument, and then the script ~/cs6038_ghidra_scripts/HelloScript.py is executed as a post-analysis script.
Many of the scripts performing work you’d typically run within the GUI using the Script Manager will be run
as post-analysis scripts. Some more advanced use cases that involve modifications to the program’s database or
other environmental modifications - where you’d like to cause Ghidra to alter the behavior of its analysis
modules - would necessitate the use of the -preScript argument instead.
Ghidra, likewise, has the ability to import other artifacts as well from the command-line. Hypothetically, were
we to have an algo2 binary, we could import it this way:
/opt/ghidra_9.1.1_PUBLIC/support/analyzeHeadless ~/gproj/ demo1 \
-import algo2
After which, we may want to do the following to run our script against it. Note that, upon import, the file is analyzed automatically. This is in contrast to the Ghidra UI, which postpones this until you open the artifact, and then prompts you:
/opt/ghidra_9.1.1_PUBLIC/support/analyzeHeadless ~/gproj/ demo1 \
-process algo2 \
-noanalysis \
-postScript ~/cs6038_ghidra_scripts/HelloScript.py
Whole Program Analysis
As mentioned earlier, the command-line analyzeHeadless doesn’t use the GUI, and therefore doesn’t have a
cursor position tracking the concept of “current function” or “current line of code”. In order to perform our
analysis, it is necessary to utilize the Ghidra environment to look up discovered functions from the analysis
DB. Borrowing the core algorithm from GetFunctionInstructions.py, we can add a loop stage to walk every
function discovered within the program, and perforom the instruction summary-count report generation on each,
wrapping this up into a single report containing all functions.
For this example, we use the Program class and its getFunctionManager() method, which provdies a
FunctionManager
instance. This can then be used to interrogate the analyzer for an inventory of functions. I then take the
single-function algorithm I wrote earlier, and have it run on every single function in the program.
GetAllFunctionsInstructions.py:
# Retrieves and displays the summary counts for instructions used within
# a function.
#
# @category CS6038.Demo
#
# Use the Python json library
import json
# Import some of the Ghidra classes we will be using
from ghidra.util.task import ConsoleTaskMonitor
# Initialize an empty dict for the "all functions" report
fn_report = {}
# the Program.getFunctionManager() provides an interface to navigate the functions
# that Ghidra has found within the program. The getFunctions() method will provide
# an iterator that allows you to walk through the list forward (True) or
# backward (False).
for fn in getCurrentProgram().getFunctionManager().getFunctions(True):
# Get the earliest instruction defined within the function, to start our exploration
instr = getInstructionAt(fn.getBody().getMinAddress())
# If it is defined, then we assume this is a navigable function and create an entry
# for it in fn_report
if instr:
fn_report[fn.getName()] = {}
# This code is largely the same as the GetFunctionInstructions.py code, with the change
# that it uses the functions provided from the aforementioned iterator, rather than the
# function from the cursor position
while instr and instr.getMinAddress() <= fn.getBody().getMaxAddress():
if fn.getBody().contains(instr.getMinAddress()):
opcode = instr.getMnemonicString()
if opcode in fn_report[fn.getName()]:
fn_report[fn.getName()][opcode] += 1
else:
fn_report[fn.getName()][opcode] = 1
instr = instr.getNext()
# Display the report to the console
print(json.dumps(fn_report, sort_keys=True, indent=2))
# The below is useful if you want to print raw JSON for machine-machine handling
#print(json.dumps(fn_report))
Running the above, you’ll likely observe that there are a bunch of messages from the Ghidra environment (which you probably saw with the Hello examples), and in the middle of that is the output containing your JSON. Below is an excerpt:
INFO HEADLESS: execution starts (HeadlessAnalyzer)
INFO Opening existing project: /home/kali/gproj/demo1 (HeadlessAnalyzer)
INFO Opening project: /home/kali/gproj/demo1 (HeadlessAnalyzer$HeadlessProject)
INFO REPORT: Processing project file: /algo1 (HeadlessAnalyzer)
WARNING: An illegal reflective access operation has occurred
WARNING: Illegal reflective access by net.sf.cglib.core.ReflectUtils$2 (file:/opt/ghidra_9.1.1_PUBLIC/Ghidra/Framework/Generic/lib/cglib-nodep-2.2.jar) to method java.lang.ClassLoader.defineClass(java.lang.String,byte[],int,int,java.security.ProtectionDomain)
WARNING: Please consider reporting this to the maintainers of net.sf.cglib.core.ReflectUtils$2
WARNING: Use --illegal-access=warn to enable warnings of further illegal reflective access operations
WARNING: All illegal access operations will be denied in a future release
INFO SCRIPT: /home/kali/src/cs6038_ghidra_scripts/GetAllFunctionsInstructions.py (HeadlessAnalyzer)
{
"FUN_00401020": {
"JMP": 1,
"PUSH": 1
},
"FUN_00401180": {
"CALL": 2,
"MOV": 6,
"POP": 1,
"PUSH": 1,
"RET": 1
},
"FUN_004011c0": {
"CALL": 2,
"MOV": 7,
"POP": 1,
"PUSH": 1,
"RET": 1
},
...
Parsing Arguments and Writing Output to a File
One of the challenges with this approach is that it is difficult to separate the JSON report output from the other reporting information. I would strongly recommend adding the capability for your script to write its output to a file. In the next example, I’ll show you how to have a filename provided on the command line and get processed as an argument by Python’s argparse.ArgumentParser (note that Ghidra still uses Python 2, as that is the version implemented by Jython).
First, while it is customary for argument names to begin with hyphens (like -o or --output), the way Java
is called from analyzeHeadless will hide such arguments from the GhidraScript when it runs. Instead, I found
I had to tell Python to use a different option prefix, so I chose + because it is fairly similar, and close to
the hyphen. The following block of code creates a new argument parser for the script based upon this specification,
and processes the Ghidra script’s arguments
(accessible from getScriptArgs())
into the traditional Python arguments result structure as documented by the above-linked argparse HOWTO documentation.
# Set up parser for the script arguments
arg_parser = ArgumentParser(description="Opcode statistical analysis", prog='script',
prefix_chars='+')
arg_parser.add_argument('+o', '++output', required=True, help='Output file for JSON')
args = arg_parser.parse_args(args=getScriptArgs())
The nice thing about doing this approach is that, though it takes slightly more work up front, adding new options becomes a simply copy-paste-revise exercise, and also it forces you to provide information that will be included in a “command-line help” documentation.
For instance, running the following:
/opt/ghidra_9.1.1_PUBLIC/support/analyzeHeadless ~/gproj demo1 -process algo1 -noanalysis \
-postScript ./GetAllFunctionsInstructions.py \
++help
Can display the following helpful output if another user, or even future you, is confused:
usage: script [+h] +o OUTPUT
Opcode statistical analysis
optional arguments:
+h, ++help show this help message and exit
+o OUTPUT, ++output OUTPUT
Output file for JSON
Next, I replace the print() call at the end of the script with the following, that writes my JSON into the file
given by the user:
# Now, open the file provided by the user, and write the JSON into it
with open(args.output, 'w') as outfile:
outfile.write(json.dumps(fn_report))
Finally, I have the following updated script (GetAllFunctionsInstructionsToFile.py):
# Retrieves and displays the summary counts for instructions used within
# a function.
#
# @category CS6038.Demo
#
# Use the Python json library
import json
# Add the Python argument parser
from argparse import ArgumentParser
# Import some of the Ghidra classes we will be using
from ghidra.util.task import ConsoleTaskMonitor
# Initialize an empty dict for the "all functions" report
fn_report = {}
# Set up parser for the script arguments
arg_parser = ArgumentParser(description="Opcode statistical analysis", prog='script',
prefix_chars='+')
arg_parser.add_argument('+o', '++output', required=True, help='Output file for JSON')
args = arg_parser.parse_args(args=getScriptArgs())
# the Program.getFunctionManager() provides an interface to navigate the functions
# that Ghidra has found within the program. The getFunctions() method will provide
# an iterator that allows you to walk through the list forward (True) or
# backward (False).
for fn in getCurrentProgram().getFunctionManager().getFunctions(True):
# Get the earliest instruction defined within the function, to start our exploration
instr = getInstructionAt(fn.getBody().getMinAddress())
# If it is defined, then we assume this is a navigable function and create an entry
# for it in fn_report
if instr:
fn_report[fn.getName()] = {}
# This code is largely the same as the GetFunctionInstructions.py code, with the change
# that it uses the functions provided from the aforementioned iterator, rather than the
# function from the cursor position
while instr and instr.getMinAddress() <= fn.getBody().getMaxAddress():
if fn.getBody().contains(instr.getMinAddress()):
opcode = instr.getMnemonicString()
if opcode in fn_report[fn.getName()]:
fn_report[fn.getName()][opcode] += 1
else:
fn_report[fn.getName()][opcode] = 1
instr = instr.getNext()
# Now, open the file provided by the user, and write the JSON into it
with open(args.output, 'w') as outfile:
outfile.write(json.dumps(fn_report))
When run with the following command line, it wrote the JSON out to algo1.json:
/opt/ghidra_9.1.1_PUBLIC/support/analyzeHeadless ~/gproj demo1 -process algo1 -noanalysis \
-postScript ./GetAllFunctionsInstructionsToFile.py \
++output algo1.json
Accessing Ghidra P-Code
As discussed earlier on, Ghidra translates the disassembly into P-Code micro-operations which are then analyzed to help decompile the code into readable C/C++. Analyzing this can provide a common language that may facilitate cross-architectural analysis that is not possible at the disassembly level.
Ghidra provides the getPcode() interface in ghidra.program.model.listing.Instruction. This outputs an array of PcodeOp instances, that represent zero or more P-Code operations that are used to represent the machine instruction in Ghidra.
Modifying the above GetAllFunctionsInstructionsToFile.py script slightly, we can create a new script
that performs roughly the same analysis for P-Code, named
GetAllFunctionsPcodeToFile.py:
# Retrieves and displays the summary counts for P-Code ops used within
# a function.
#
# @category CS6038.Demo
#
# Use the Python json library
import json
# Add the Python argument parser
from argparse import ArgumentParser
# Import some of the Ghidra classes we will be using
from ghidra.util.task import ConsoleTaskMonitor
# Initialize an empty dict for the "all functions" report
fn_report = {}
# Set up parser for the script arguments
arg_parser = ArgumentParser(description="P-Code statistical analysis", prog='script',
prefix_chars='+')
arg_parser.add_argument('+o', '++output', required=True, help='Output file for JSON')
args = arg_parser.parse_args(args=getScriptArgs())
# the Program.getFunctionManager() provides an interface to navigate the functions
# that Ghidra has found within the program. The getFunctions() method will provide
# an iterator that allows you to walk through the list forward (True) or
# backward (False).
for fn in getCurrentProgram().getFunctionManager().getFunctions(True):
# Get the earliest instruction defined within the function, to start our exploration
instr = getInstructionAt(fn.getBody().getMinAddress())
# Walk through each instruction that's determined to be part of this function
while instr and instr.getMinAddress() <= fn.getBody().getMaxAddress():
if fn.getBody().contains(instr.getMinAddress()):
# Iterate across the list of P-Code operations that are expanded from
# the parsed machine instruction
for pcode_op in instr.getPcode():
# Get the string name of the PCode operation
pcode_name = pcode_op.getMnemonic()
# Create a new report for this function the first time we get a valid instruction
if fn.getName() not in fn_report:
fn_report[fn.getName()] = {}
if pcode_name in fn_report[fn.getName()]:
fn_report[fn.getName()][pcode_name] += 1
else:
fn_report[fn.getName()][pcode_name] = 1
# Advance to the next instruction
instr = instr.getNext()
# Now, open the file provided by the user, and write the JSON into it
with open(args.output, 'w') as outfile:
outfile.write(json.dumps(fn_report))
Similar to earlier, I can execute this script to generate algo1_pc.json:
/opt/ghidra_9.1.1_PUBLIC/support/analyzeHeadless ~/gproj demo1 -process algo1 -noanalysis \
-postScript ./GetAllFunctionsPcodeToFile.py \
++output algo1_pc.json
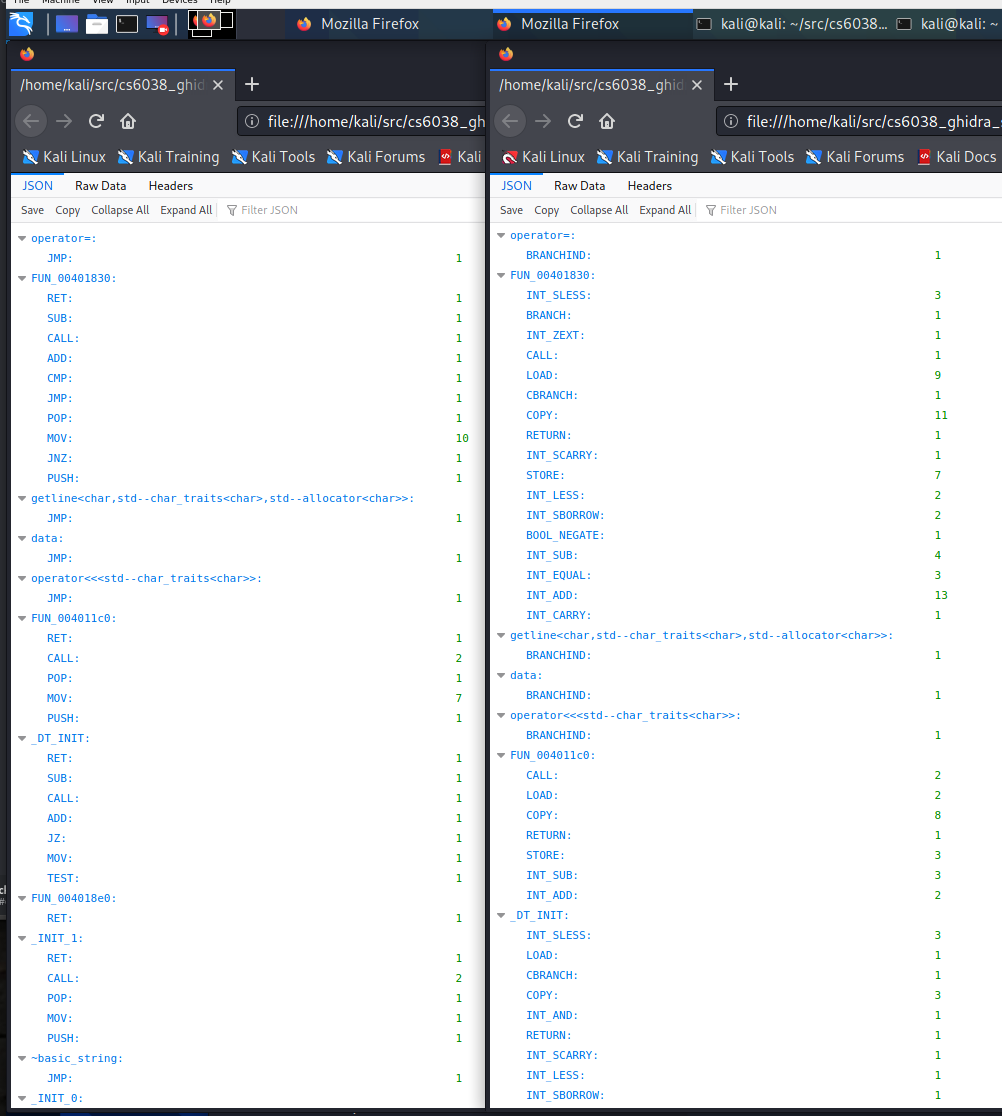
In my case, I ran both of these from teh same folder to generate both JSON files. I can view these with a JSON viewer, such as the Firefox web browser, or even the command-line utility jq. Firefox is nice and easy and already installed on the Kali VM provided for class:
firefox algo1*.json
Firefox brought up both files in separate tabs, pulling one out into its own window and looking side
by side at the same function in both views yields some nice insights: